Portfolio
Here I am slowly collecting / presenting work that I have done. While I am constructing this simplified overview, please also have a look at my CV and my scientific output. The corresponding repository with, in some cases, more info is at GitHub.
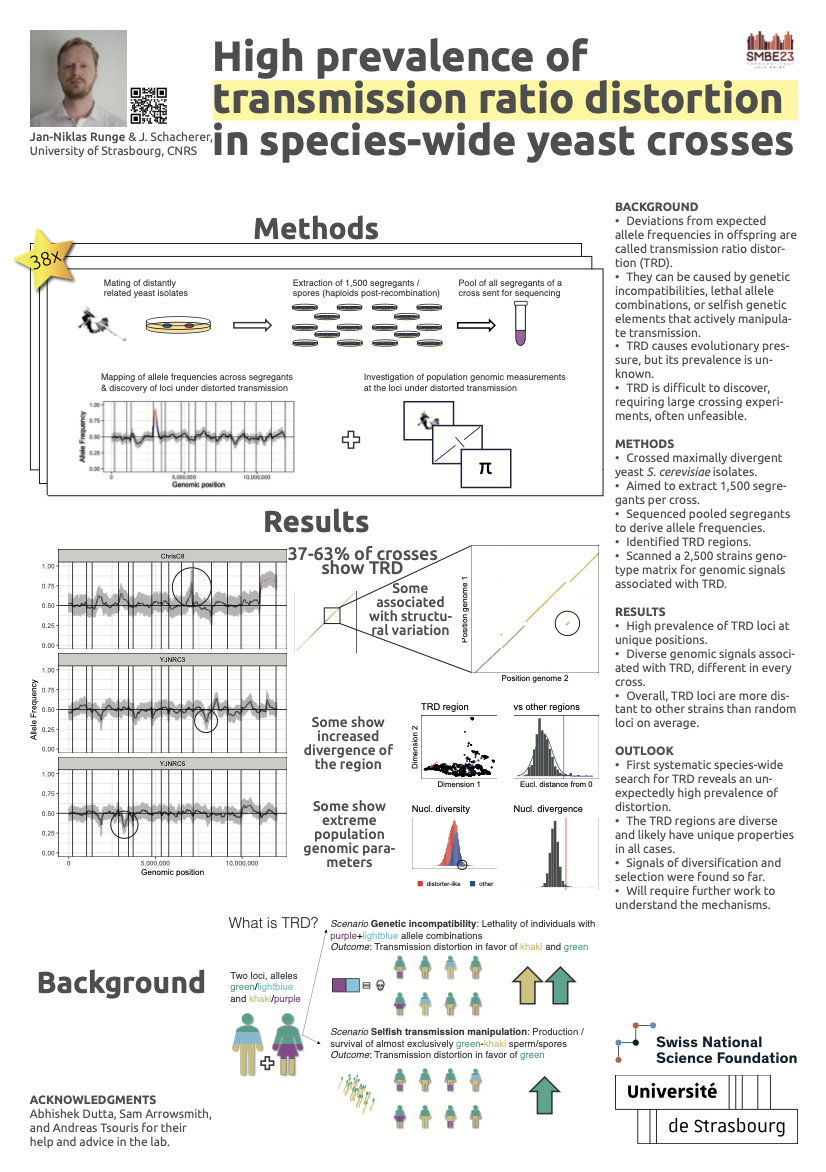
Transmission ratio distortion project
In this currently ongoing project I aim to find genomic signals underlying loci under distorted transmission. To that end, I went to the wet lab in Strasbourg to cross distantly related yeast isolates, retrieve their haploid offspring, pool them, and sequence them to extract genome-wide allele frequencies. This allows me to quantify the prevalence of transmission distortion across a species, something that came up as a question over and over again in talks I gave on meiotic drive in house mice. Thanks to the great resources at the Schacherer Lab, I am able to then take those distorted loci and see if any interesting, and in part hypothesized, genomic signals, such as changes in local phylogeny, nucleotide diversity, or structural variation are associated with distortion! A preview of what that looks like can be seen in the poster right next to this text, which I presented at SMBE 2023.
Work in progress repository: GitHub
Assembly of a selfish haplotype in house mice
We are currently finishing this project up and will have more to share after.
Short video summary
Investigating inbreeding in a long-term population
We began working on the causes and consequences of inbreeding in the long-term study population after we discovered the high levels of inbreeding in that population. Some of the preliminary work can be seen in the video summary.
Short video summary
Parent-offspring inference in inbred populations
When we began working on the genotypes of the long-term study population of house mice, we quickly found that traditional pedigree (i.e. relationships between mice) inference methods were not working. The high levels of inbreeding obfuscated genetic relationships so much that most tools would infer every mouse to be siblings. Because of that, we had to develop our own approach to pedigree inference, which we validated and published.
SPORE can be found at GitHub and the paper can be found at Molecular Ecology Resources.
Short video summary
The genotype imputation pipeline for a 10,000+ mouse genomics project
To prepare for future analyses, I became the person responsible for generating whole-genome genotypes from ultra-low coverage sequencing of thousands of wild house mouse genomes. It was an awesome challenge at the time and I used a variety of R, bash, and Python scripts on a high performance cluster over many cycles of new data and updates to the pipeline to get to a really cost-effective (in terms of sequencing cost) outcome, which will be used for years to come.
An outdated and incomplete snapshot of this pipeline can be found at GitHub while I am finishing it up for the actual repository.
Short video summary
Sequencing progress dashboard
For my work at Columbia University, where I was responsible for the full data cycle from raw sequencing data to imputed genotypes, the inferred pedigree, etc., for 10,000+ mice, computational times were enormous. I needed a quick overview of where things are at and for this purpose, I quickly put together a dashboard in R Shiny that can be run from anyone with server access to check the progress. In the background, it logs onto the server, fetches files that I am generating as part of the genotyping pipeline, and presents them in an intuitive overview.
Anonymized source code: GitHub
Short video of the app's output
Simulations of phenotype evolution of a selfish genetic element
For my third PhD paper, I came up with the idea to improve our understanding of the phenotypic impact of the selfish genetic element by simulating the evolution of this trait. To that end, I learned to write custom agent-based models (built on NetLogo) and over several iterations always kept finding the same result: Yes, this phenotypic difference should evolve! The analysis of this data and the execution was the first real big data challenge for me at the time.
The paper can be found here: Journal of Evolutionary Biology
Short video summary
Experimental validation of a newly discovered behavioural phenotypic difference between genotypes
For my second PhD paper, I built experimental setups to test for differences in several behaviours associated with a hypothesized "dispersal syndrome", which is a collection of behaviours thought to be changed in individuals that are more likely to disperse / emigrate from populations. I used scored observations of video recordings of exploration experiments to extract several different variables describing the mice's behaviour. I condensed these correlated variables down using a PCA and found differences between genotypes in the main dimension of the PCA. I also analysed emigration probabilities from the experimental setup over time using Cox survival models to validate the increased emigration propensity of the genotypes.
The paper can be found here: Royal Society Open Science
Short video summary
Discovery of a behavioural phenotype of a selfish genetic element via long-term data analysis
For my very first PhD paper, I employed generalized linear mixed models to understand emigration (or dispersal) behaviour in a long-term wild house mouse study population. To that end, I cleaned the data, and inferred emigration behaviour based on disappearances of young mice from the population. After building a null model of emigration based on relevant individual and environmental determinants (sex, population density, and year of birth), I tested whether adding information on a specific genotype (a selfish genetic element) could explain some of the individual differences in emigration propensity. Indeed, using bootstrapping, I found this to be strongly the case, with almost 50% increased odds at average densities.
The paper can be found here: Proceedings of the Royal Society B